Biomedical Network Science Lab
The Biomedical Network Science (BIONETS) Lab investigates molecular mechanisms, using techniques from network science, (graph-based) AI and combinatorial optimization. We develop algorithms, AI models, and software tools to mine omics data for such mechanisms, with the aim to better understand cellular pathways and, ultimately, pave the way for targeted and causally effective treatments of complex diseases. We also develop privacy-preserving decentralized biomedical AI solutions, which enable cross-institutional studies on sensitive data. Finally, we are interested in meta-scientific questions such as reproducibility and the impact of data bias on biomedical AI systems.

News
AIBeez
We are very happy to announce the birth of AIBeez – “Association for the Promotion of Artificial Intelligence in Health and Engineering Sciences in Erlangen e.V.” New members who want to support AI research for biomedicine in Erlangen are very welcome!
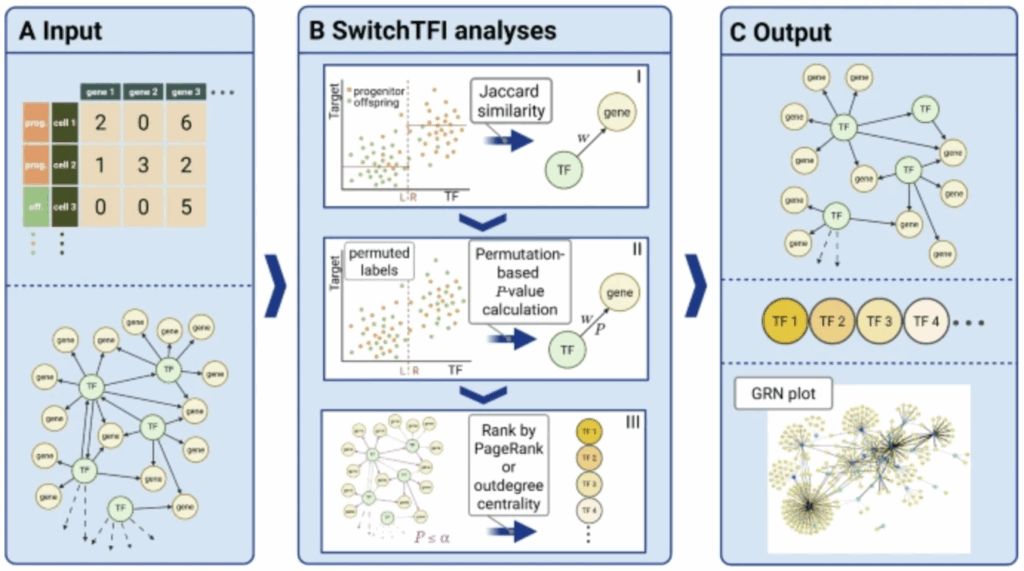
Paper published in Genome Biology
In our paper “SwitchTFI: identifying transcription factors driving cell differentiation”, we present an algorithm to infer regulatory mechanisms that drive the transition from progenitor to offspring cells from scRNA-seq data.
DFG funds CASCAID
We are very excited to announce that the DFG funds the new CRC “Cellular and Systems Control of Autoimmune Disease” (CASCAID). Together with the lab of Stefan Uderhardt, we will develop graph-based models of histological healing that quantify the target of successful treatment of autoimmune diseases. Moreover, we will provide CASCAID-wide bioinformatics and data analysis…
Paper published in GigaScience
In our paper “NApy: Efficient Statistics in Python for Large-Scale Heterogeneous Data with Enhanced Support for Missing Data”, we present a Python package with Numba and OpenMP-powered C++ backends for fast and memory-efficient statistical testing in the presence of missing data.
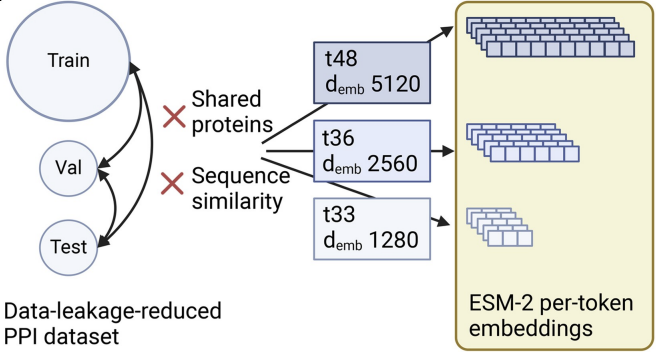
Paper published in ISMB 2025 proceedings
In our paper “Deep learning models for unbiased sequence-based PPI prediction plateau at an accuracy of 0.65” we show that usage of ESM-2 embeddings boosts performance in out-of-distribution PPI prediction to around 0.65 independently of model architecture.